Monday, February 23, 2009 / Labels: Biology Molecular
Molecular Farming
Plants do not carry pathogens that might be dangerous to human health. Additionally, on the level of pharmacologically active proteins, there are no proteins in plants that are similar to human proteins. On the other hand, plants are still sufficiently closely related to animals and humans that they are able to correctly process and configure both animal and human proteins. Their seeds and fruits also provide sterile packaging containers for the valuable therapeutics and guarantee a certain storage life.
Global demand for pharmaceuticals is at unprecedented levels, and current production capacity will soon be overwhelmed. Expanding the existing microbial systems, although feasible for some therapeutic products, is not a satisfactory option on several grounds. First, it would be very expensive for the pharmaceutical companies. Second, other proteins of interest are too complex to be made by microbial systems. These proteins are currently being produced in animal cell cultures, but the resulting product is often prohibitively expensive for many patients. Finally, although it is theoretically possible to synthesize protein molecules by machine, this works only for very small molecules, less than 30 amino acid residue in length. Virtually all proteins of therapeutic value are larger than this and require live cells to produce them. For these reasons, science has been exploring other options for producing proteins of therapeutic value.

While molecular farming is one application of genetic engineering, there are concerns that are unique to it. In the case of genetically modified (GM) foods, concerns focus on the safety of the food for human consumption. In response, it has been argued that the genes that enhance a crop in some way, such as drought resistance or pesticide resistance, are not believed to affect the food itself. Other GM foods in development, such as fruits designed to ripen faster or grow larger, are believed not to affect humans any differently from non-GM varieties. In contrast, molecular farming is not intended for crops destined for the food chain. It produces plants that contain physiologically active compounds that accumulate in the plant’s tissues. Considerable attention is focussed, therefore, on the restraint and caution necessary to protect both consumer health and environmental biodiversity.
There are also problems associated with the use of plants as protein bioreactors. Plant proteins have different sugar residues from human or animal proteins. Freiburg-based greenovation Biotech GmbH, in cooperation with Professor Ralf Reski’s research group at the University of Freiburg, has shown that this problem can be solved through the use of Physcomitrella patens. Because the scientists cultivate the moss in tube-shaped photobioreactors in a liquid medium, they have no worries that the genetically modified mosses might be released into the environment.
Tuesday, February 17, 2009 / Labels: Biology Molecular
Natural Product compound
Natural products have long been a major source of new medicines. About 50% of the drugs introduced to the market in the last 20 years trace their origin to compounds derived from nature. A similar impact has been experienced in animal health and agrochemical discovery.

Introduction
Before the 20th century, crude and semi-pure extracts of plants, animals, microbes and minerals represented the only medications available to treat human and domestic animal illnesses. The 20th century revolutionized the thinking in the use of drugs, as the receptor theory of drug action was postulated in ?. The idea that effect of drug in human body are mediated by specific interactions of the drug molecule with biological macromolecules, (proteins or nucleic acids in most cases) led scientists to the conclusion that individual chemical compounds in extracts, rather than some mystical “power of life” are the factors required for the biological activity of the drug. This made for the beginning of a totally new era in pharmacology, as pure, isolated chemicals, instead of extracts, became the standard treatments for diseases. Indeed, many bioactive compounds, responsible for the effects of crude extract drugs, and their chemical structure was elucidated. Classical examples of drug compounds discovered this way are morphine, the active agent in opium, and digoxin, a heart stimulant originating from flower Digitalis lanata. The evolution in synthetic chemistry also led to chemical synthesis of many of the elucidated structures.
Indeed, the 20th century brought up several new drug compounds, and until the 1970s, the new drug compounds were almost solely of natural origin. However, as the fields of synthetic chemistry became more and more powerful, the pharmaceutical industry started to prefer synthetic compounds instead of natural products as drug candidates. The following reasons for the decline in interest in natural products as drug candidates have been suggested: [1]
introduction of high-throughput screening (HTS) as the standard method for hit discovery. The traditional natural product libraries were poorly suitable for HTS environment.
the pressure to faster generation of lead compounds. The process in natural product drug discovery usually required several separation circles and structure elucidation (see below) and was thus time-consuming.
rise of combinatorial chemistry and thus the generation of synthetic compound libraries in a screening friendly format
general decline in interest towards developing new antibiotic drugs, a traditionally strong area of natural product drug discovery.
However, more recent evolvements in techniques involved in natural product research, as well as the observation of the chemical complementarity of natural and synthetic compounds, have restored the interest in natural compounds as drug candidates. The declining trend in patents on natural products has turned as a slight increase in the beginning of the 21st century.
Nature as source of drug compounds
Despite the rise of combinatorial chemistry as an integral part of lead discovery process, the natural products still play a major role as starting material for drug discovery.[2] David Newman and Gordon Cragg have made a remarkable contribution to evaluation of the significance of natural products in drug discovery via their analysis of the sources of approved drugs. The latest update of the report was published in 2007 [3], covering years 1981-2006. According to their report, of the 974 small molecule new chemical entities, 63% were natural derived or nature-inspired (semisynthetic derivatives of natural products, compounds synthesised by use of natural product pharmacophore or compounds otherwise designed to mimic the natural ligand/substrate of the target). For certain therapy areas, such as antimicrobials, anticancer antihypertensive and anti-inflammatory drugs, the numbers were even higher (for instance, approximately 75% of all approved small molecule new chemical entities were derived from nature.
Natural products have been especially successful as lead structures for antibacterial therapies
A potential explanation beyond the success of natural products as drugs is the classification of natural compounds as so-called privileged structures. This is because chemical agents produced by living organisms (particularly the secondary metabolites) have evolved over millennia under the evolutionary pressure, and are therefore more likely to have a specific biological activity than “randomly” assembled, man-made synthetic chemicals. Despite the enormous potential, only a minor fraction of globe’s living species has ever been tested for any bioactivity. For instance, approximately only 10% of all existing plant species has been assayed, and in the case of microbes the value is even lower.
Plant-derived bioactive material
The vast majority of traditionally used crude drugs have been plant-derived extracts. This has resulted in an inherited pool of information of the healing potential of plant species, thus making them important source of starting material for drug discovery. A different set of metabolites is usually produced in the different anatomical parts of the plant (e.. root, leaves and flower), and botanical knowledge is crucial also for the correct taxonomical determination of the identified bioactive plants.
Microbial species with bioactive metabolites
In the microbial world, there is an ongoing, everlasting competition of living space and nutrients. To survive in these conditions, many microbes have developed abilities to prevent competing species from proliferation. This phenomenon has been translated to the introduction of microbes as the main source of antimicrobial drugs, even though some of these secondary metabolites have also other potent biological activities as well. For the antibacterials, different Streptomyces species have been the most productive bacteria. The classical example of an antibiotic discovered as a defense mechanism against another microbe is the discovery of penicillin in the cultures of Penicillum fungi in 1928.
Marine invertebrates as a source for bioactive compounds
Besides terrestrial ecosystems, marine environments are considered potential sources for new bioactive agents. The first breakthroughs in the area were the arabinose nucleosides discovered from marine invertebates in 1950s, demonstrating for the first time that also sugar moieties other than ribose and deoxyribose can yield bioactive nucleoside structures. However, it took as long as 2004 until the first marine-derived drug was approved. The cone snail toxin ziconotide, also known as Prialt, was then approved by Food and Drug Administration (FDA, USA) to treat severe neuropathic pain. Several other marine-derived agents are now in clinical trials for indications such as cancer, anti-inflammatory use and pain. One of the most promising classes of these agents in pipeline are bryostatin-like compounds, that are under investigation as anti-cancer therapy as such and particularly as combination with other cytostatic drugs.
Chemical diversity of Natural Products
As above mentioned, combinatorial chemistry was a key technology enabling the efficient generation of large screening libraries for the needs of high-throughput screening. However, now, after two decades of combinatorial chemistry, it has been pointed out that despite the increased efficiency in chemical synthesis, no increase in lead or drug candidates has been reached [2]. This has led to analysis of chemical characteristics of combinatorial chemistry products, compared to existing drugs and/or natural products. The chemoinformatics concept chemical diversity, depicted as distribution of compounds in the chemical space based on their physicochemical characteristics, is often used to describe the difference between the combinatorial chemistry libraries and natural products. The synthetic, combinatorial library compounds seem to cover only a limited and quite uniform chemical space, whereas existing drugs and particularly natural products, exhibit much greater chemical diversity, distributing more evenly to the chemical space. The most prominent differences between natural products and compounds in combinatorial chemistry libraries is the number of chiral centers (much higher in natural compounds), structure rigidity (higher in natural compounds) and number of aromatic moieties (higher in combinatorial chemistry libraries). Other chemical differences between these two groups include the nature of heteroatoms (O and N enriched in natural products, and S and halogen atoms more often present in synthetic compounds), as well as level of non-aromatic unsaturation (higher in natural products). As both structure rigidity and chirality are both well-established factors in medicinal chemistry known to enhance compounds specificity and efficacy as a drug, it has been suggested that natural products compare favourable to today's combinatorial chemistry libraries as potential lead molecules.
Methodologies in natural product drug discovery
Identification of biologically active material
Two main approaches exist for the finding of new bioactive chemical entities from natural sources; either random collection and screening of material, or exploitation of ethnopharmacological knowledge in the selection. The former approach bases itself on the fact that only a very small part of globes’s biodiversity has ever been tested for any biological activity, and on the other hand, particularly organisms living in a species-rich environment need to evolve defence and competition mechanism to survive. Thus, collection of plant, animal and microbial samples from rich ecosystems may give rise to isolation of novel biological activities. One example of a successful use of this strategy is the screening for antitumour agents, performed by National Cancer Institute in USA started in 1960s. Cytostate paclitaxel (taxoid) was identidifed during this campaign from Pacific yew tree Taxus brevifolia. Paclitaxel showed anti-tumour activity with previously unknown mechanism (stabilization of microtubules) and is now approved for clinical use for the treatment of lung, breast and ovary cancer, as well as for Kapos sarcoma.
Besides random selection, the selection of starting material may be done by collecting knowledge on use of plants and other natural products as herbal medicines and thereby get an idea of potential biological activities. Ethnobotany, the study of the use of plants in the society, and particularly ethnopharmacology, an area inside ethnobotany focused on mediical use of plants, may therefore provide invaluable information, as illustrated by the example of artemisinin, an antimalarial agent from sweet wormtree Artemisiae annua, used in Chinese medicine since 200 DC and nowadays in use against multiresistant malarial protozoa Plasmodium falciparum.
Structural elucidation
The elucidation of the chemical structure is critical to avoid double hits (i.e. identification of a chemical agent that is already known for its structure and chemical activity), and for a long time remained the most time-consuming step in natural product drug discovery. New methods have been applied in this field, thus making the task easier and faster. In particular, mass spectrometry (MS) has contributed to the enhanced ease of structure determination. MS is a method in which individual compounds are identified based on their mass/charge ratio, after an artificial ionization. Natural compounds mainly exist as mixtures (when extracted from their origin) so the combination of liquid chromatography and mass spectrometry (LC-MS) is often used to separate the individual compounds and determine their mass/charge ratios online. Databases of mass spectras for known natural compounds are available, allowing the comparisons. Besides MS, nuclear magnetic resonance (NMR) spectroscopy is another important technique for determining chemical structures of natural products. NMR yields information about individual hydrogen and carbon atoms in the structure, allowing detailed reconstruction of the molecule’s architecture.
(from wikipedia)
Wednesday, February 4, 2009 / Labels: Genetic Engineering
Genetic engineering
 Would you want to clone your pet? Would you change your child's eye color? Do you care if your strawberry contains a gene for fish?
Would you want to clone your pet? Would you change your child's eye color? Do you care if your strawberry contains a gene for fish?Genetic Engineering tells you the story, gives you the facts, and then takes a closer look to help you unravel the core issues. Take a look at and interact with the content. Discuss what you learn with other people, form your own opinion on the subjects, but always keep an open mind.
Genetic engineering is a laboratory technique used by scientists to change the DNA of living organisms.
DNA is the blueprint for the individuality of an organism. The organism relies upon the information stored in its DNA for the management of every biochemical process. The life, growth and unique features of the organism depend on its DNA. The segments of DNA which have been associated with specific features or functions of an organism are called genes.
Molecular biologists have discovered many enzymes which change the structure of DNA in living organisms. Some of these enzymes can cut and join strands of DNA. Using such enzymes, scientists learned to cut specific genes from DNA and to build customized DNA using these genes. They also learned about vectors, strands of DNA such as viruses, which can infect a cell and insert themselves into its DNA.
With this knowledge, scientists started to build vectors which incorporated genes of their choosing and used the new vectors to insert these genes into the DNA of living organisms. Genetic engineers believe they can improve the foods we eat by doing this. For example, tomatoes are sensitive to frost. This shortens their growing season. Fish, on the other hand, survive in very cold water. Scientists identified a particular gene which enables a flounder to resist cold and used the technology of genetic engineering to insert this 'anti-freeze' gene into a tomato. This makes it possible to extend the growing season of the tomato.
Isolation of the genes of interest
Insertion of the genes into a transfer vector
Transfer of the vector to the organism to be modified
Transformation of the cells of the organism
Selection of the genetically modified organism (GMO) from those that have not been successfully modified
Thursday, January 29, 2009 / Labels: Gene Expression
Protein expression
 Protein expression is a subcomponent of gene expression. It consists of the stages after DNA has been translated into amino acids chains, which are ultimately folded into protein. Protein expression is commonly used by proteomics researchers to denote the measurement of the presence and abundance of one or more proteins in a particular cell or tissue.
Protein expression is a subcomponent of gene expression. It consists of the stages after DNA has been translated into amino acids chains, which are ultimately folded into protein. Protein expression is commonly used by proteomics researchers to denote the measurement of the presence and abundance of one or more proteins in a particular cell or tissue.Protein expression systems include bacterial, yeast, baculovirus/insect, and mammalian expression systems.
E. coli Expression System
Escherichia coli (E. coli) is one of the most widely used hosts for the production of heterologous proteins and its genetics are far better characterized than those of any other microorganism. Recent progress in the fundamental understanding of transcription, translation, and protein folding in E. coli, together with serendipitous discoveries and the availability of improved genetic tools are making this bacterium more valuable than ever for the expression of complex eukaryotic proteins.
Yeast Expression System
Yeast is a eukaryotic organism and has some advantages and disadvantages for protein expression as compared to E. coli. One of the major advantages is that yeast cultures can be grown to very high densities, which makes them especially useful for the production of isotope labeled protein for NMR. The two most used yeast strains are Saccharomyces cerevisiae and the methylotrophic yeast Pichia pastoris. Various yeast species have proven to be extremely useful for expression and analysis of eukaryotic proteins. These yeast strains have been genetically well characterized and are known to perform many posttranslational modifications. These single-celled eukaryotic organisms grow quickly in defined medium, are easier and less expensive to work with than insect or mammalian cells, and are easily adapted to fermentation. Yeast expression systems are ideally suited for large-scale production of recombinant eukaryotic proteins.
In some instances the most cost-effective expression of functional enzymes is the yeast expression system.
Insect Cell Expression System
Insect cells are a higher eukaryotic system than yeast and are able to carry out more complex post-translational modifications than the other two systems. They also have the best machinery for the folding of mammalian proteins and, therefore, give you the best chance of obtaining soluble protein when you want to express a protein of mammalian origin. The most commonly used vector system for recombinant protein expression in insect is baculovirus, although baculoviral also can be used for gene transfer and expression in mammalian cells
Mammalian Expression System
The production of proteins in mammalian cells is an important tool in numerous scientific and commercial areas. For example, the proteins expressed in and purified from mammalian cell system are routinely needed for life science research and development. In the field of biomedicine, proteins for human therapy, vaccination or diagnostic applications are typically produced in mammalian cells. Gene cloning, protein engineering, biochemical and biophysical characterization of proteins also require the use of gene expression in mammalian cells. Other applications in widespread use involve screening of libraries of chemical compounds in drug discovery, and the development of cell-based biosensors.
Tuesday, January 6, 2009 / Labels: Gene Expression
Gene Expression
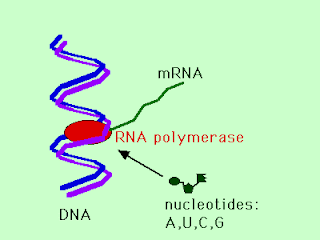
Transcription is the transfer of the genetic information from the archival copy of DNA to the short-lived messenger RNA. The enzyme RNA polymerase binds to a particular region of the DNA and starts to make a strand of mRNA wiht a base sequence complementary to the DNA template that is "downstream" of the RNA polymerase binding site. When this transcription is finished, the portion of the DNA that coded for a protein, i.e. a gene, is now represented by a messenger RNA molecule that can be used as a template for translation.
The steps in transcription are:
1.DNA unzips and RNA polymerase enzyme binds to one strand of DNA
2.RNA polymerase makes an elongating chain of RNA nucleotides, each new RNA nucleotide complementary to the DNA nucleotide it is hydrogen bonded to
3.The completed mRNA molecule is released from RNA polymerase - DNA complex and can begin translation. In eukaryotic cells this means first moving from the nucleus into the cytoplasm. In prokaryotic cells (bacteria), ribosomes can bind and begin translation before polymerase has completed of the new mRNA strand.
Translation is the process that takes the information passed from DNA as messenger RNA and turns this into a series of amino acids bound together with peptide bonds. It really is a translation from one code, nucleotide sequence, to another code, amino acid sequence. The ribosome is the site of this action, just as RNA polymerase was the site of mRNA synthesis. The ribosome matches the base sequence on the mRNA in sets of three bases (called codons) to tRNA molecules that have the three complementary bases in their anticodon regions. Again, the base pairing rule is important in this recognition (A binds to U and C binds to G). The ribosome moves along the mRNA, matching 3 base pairs at a time and adding the amino acids to the polypeptide chain. When the ribosome reaches one of the "stop" codes, the ribosome releases both the polypeptide and the mRNA. This polypeptide will twist into its native coformation and begin to act as a protein in the cells metabolism. This may be a binding protein, an enzyme, a membrane channel or transport site, or part of the electron transport chain. This description is for the simplest case such as some examples of bacterial protein synthesis. Eukaryotic cells follow these steps but other control steps and modifications are common.
The steps in translation are:
the ribosome binds to mRNA at a specific area
the ribosome starts matching tRNA anticodon sequences to the mRNA codon sequence
each time a new tRNA comes into the ribosome, the amino acid that it was carrying gets added to the elongating polypeptide chain
the ribosome continues until it hits a stop sequence, then it releases the polypeptide and the mRNA
the polypeptide forms into its native shape and starts acting as a functional protein in the cell
Tuesday, December 23, 2008 / Labels: Biology Molecular
Cell Biology
We may distinguish two general types of cells:
- Bacteria are the Prokaryotes, organisms in which nominally there is only a single cell compartment, bounded by a membrane or membranes that give security against the outside world ( Prokaryote cells DO NOT have a cell membrane around their nucleus). There are two kinds of prokaryotes, bacteria and archaea, but these are similar in the overall structures of their cells

- Eukaryotes are defined by the division of each cell into a nucleus that contains the genetic material, surrounded by a cytoplasm which in turn is bounded by the plasma membrane that marks the periphery of the cell. The cytoplasm contains other discrete compartments, also bounded by membranes. Eukaryotic cells usually are 10 times larger than Prokaryotic cells.
 These two types of cells are structurally very different. A major structural difference between prokaryotes and eukaryotes, other than size is the arrangement of DNA within the cell. Prokaryotic cells are usually independent, while eukaryotic cells are often found in multicellular organisms.
These two types of cells are structurally very different. A major structural difference between prokaryotes and eukaryotes, other than size is the arrangement of DNA within the cell. Prokaryotic cells are usually independent, while eukaryotic cells are often found in multicellular organisms.
References:
- Garrett and Grisham. BIOCHEMISTRY. Saunders College Publishing. Harcourt Brace College Publishers
- M.T. Madigan et al. Biology of Microorganisms. Prentice Hall. International Inc